Abstract
Chest X-rays are among the most commonly ordered imaging tests. Applying deep learning techniques to X-ray images is a typical application of computer vision in healthcare. Nevertheless, using X-ray images alone does not always lead to generalizable model performances. Combining patients’ clinical reports, which contain rich and important patient diagnostic information, with X-ray images could give the model more information for prediction. Consequently, this project will be focusing on deriving and examining the best fusion strategies for implementing a multimodal approach with regard to pleural effusion prediction. Using X-ray images and clinical text reports, we mainly combined VGG16 with DistilBERT to better predict the presence of pleural effusion. Two sets of fusion strategies are proposed, namely early fusion, where we concatenate the learned vector representations of images and texts before classification, and late fusion, where we leverage the predicted probabilities from the two modalities. Ultimately, we found that the late fusion multimodality model with an elastic net regularized logistic regression model achieved the best overall performance, with an AUC value of 0.9887. On the other hand, the early fusion strategy achieved inferior results, which indicates that the early fusion strategy that we utilized here is not specifically suitable for integrating X-ray images with clinical text data.
FYI:
-
The manuscript for this project can be accessed through this link.
-
You can also click this link to check out the short presentation slides that I made for this project.
Data
- MIMIC-CXR Database v2.0.0
- X-ray images in DICOM files
- Contains clinical reports
- MIMIC-CXR-JPG Database v2.0.0
- X-ray images in JPG files
- 5.5 TB in total size
- Due to the large size of the files and limitations of computing power, we eventually only used about 10% of the entire dataset
Results
For our baseline imaging model, the VGG16-based CNN with data augmentations, it ultimately achieved a test accuracy of 84.92% and an AUC of 0.9099. Nevertheless, this model consistently overfitted on the training data, regardless of the application of data augmentation techniques, as shown in the Figure below. Consequently, this indicates that this model lacks a good generalizability to unseen data beyond the overfitting point.
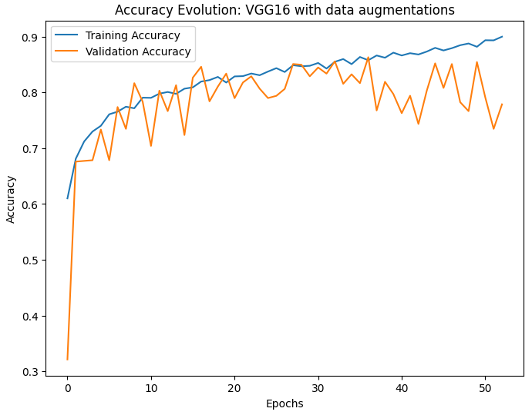
Furthermore, for our early fusion approach towards our multimodality model, the validation accuracy appeared to plateau at around 81%, even under elongated training time, as shown in Figure 1b. This suggests that the early fusion strategy may not be fully capitalizing on the potential synergies between the features of the X-ray images and the clinical texts to maximize the predictive accuracy. Ultimately, this early fusion strategy achieved an AUC of 0.8791, which is slightly worse than that of our baseline model.
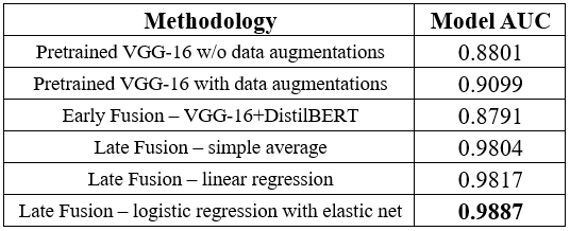
Our late fusion strategy achieved much more success compared to the aforementioned baseline approach and the early fusion multimodality model. To elaborate, a simple averaging of the predicted probability values from both models resulted in an AUC of 0.9804. For the regression-based late fusion approaches, by implementing a linear regression using the predicted probabilities, we were able to achieve an AUC of 0.9817. Remarkably, when we applied a logistic regression model with elastic net regularization, the AUC further increased to 0.9887, which is also our highest AUC value among all approaches and models. These results underline the effectiveness of late fusion multimodal learning in harnessing the predictive potential of both X-ray images and clinical reports for superior model performances.
Here is a summary of all the results mentioned above:
Lastly, to interpret our vision network, we employed two gradient-based methods: saliency maps and GradCAM, shown in the Figure below. Both techniques utilize gradients to identify important regions within an image. Saliency maps calculate the gradient of the network’s loss with respect to the input image and visualize these gradients as a heatmap (Simonyan et al., 2013). GradCAM, on the other hand, calculates gradients only for the last convolutional layer before the global pooling operation, resulting in a more abstract representation of input-output relationships and providing a different perspective on the network’s underlying mechanisms (Selvaraju et al., 2017).
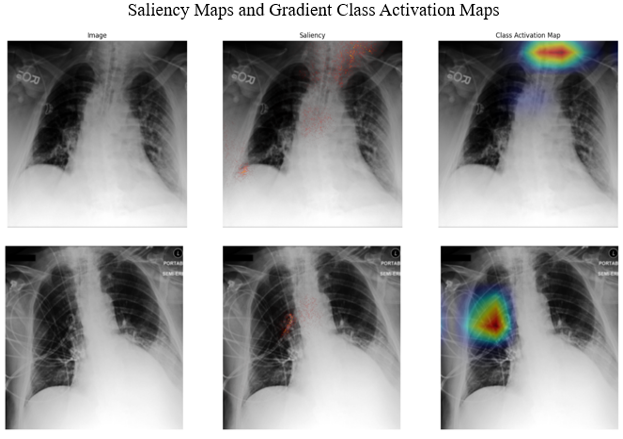
Both methods above highlighted similar regions. In addition, aside from the lung areas, the model sometimes pays attention to other regions as well as medical devices. While this could be advantageous for diagnosis, it is not ideal for model interpretability because the model’s focus is not solely on disease locations and related pathologies.
Conclusion
Overall, this project demonstrated two multimodal fusion strategies that can integrate clinical text data with X-ray image data for pleural effusion prediction. We compared their performances with our baseline VGG16 model and found that the late fusion multimodality model with an elastic net regularized logistic regression model has the best overall performance, achieving an AUC score of 0.9887. On the other hand, the early fusion strategy that concatenates learned X-ray image features and clinical text features only achieved slightly worse results than our baseline model did. Therefore, we can infer that the early fusion strategy that we utilized here is not suitable for integrating X-ray images with clinical text data. Consequently, we have shown that combining textual data with imaging data in clinical use could aid disease prediction, but to achieve better results using an early fusion architecture, a more complex and robust architecture will be needed.
Future study can work on improving early fusion architectures by identifying the most effective representations of text data and how the clinical text information can be integrated with X-ray images through the interaction and consensus between them. Furthermore, the generalizability of such models is unsure and remains an open problem since this project has only shown that clinical text can be supportive to the analysis of imaging data that are from the same database.
References
Johnson, A., Lungren, M., Peng, Y., Lu, Z., Mark, R., Berkowitz, S., & Horng, S. (2019). MIMIC-CXR-JPG - chest radiographs with structured labels (version 2.0.0). PhysioNet. https://doi.org/10.13026/8360-t248.
Sanh, V., Debut, L., Chaumond, J., & Wolf, T. (2019). DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. arXiv preprint arXiv:1910.01108.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2017). Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision (pp. 618-626).
Simonyan, K., Vedaldi, A., & Zisserman, A. (2013). Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034.